Amplicon sequence variants and bias with Benjamin Callahan (#39)
November 29, 2019
In this episode, Benjamin Callahan talks about some of the issues faced by microbiologists when conducting amplicon sequencing and metagenomic studies. The two main themes are:
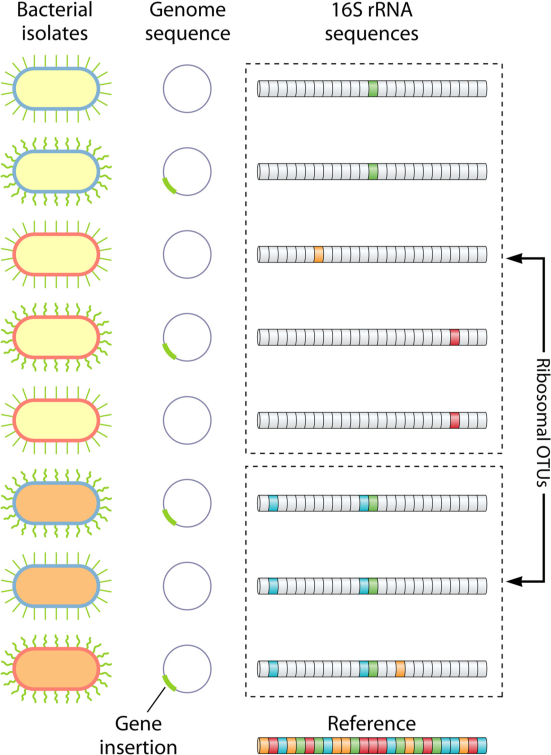
- Why one should probably avoid using OTUs (operational taxonomic units) and use exact sequence variants (also called amplicon sequence variants, or ASVs), and how DADA2 manages to deduce the exact sequences present in the sample.
- Why abundances inferred from community sequencing data are biased, and how we can model and correct this bias.
Links:
- Exact sequence variants should replace operational taxonomic units in marker-gene data analysis (Benjamin J Callahan, Paul J McMurdie & Susan P Holmes)
- DADA2: High-resolution sample inference from Illumina amplicon data (Benjamin J Callahan, Paul J McMurdie, Michael J Rosen, Andrew W Han, Amy Jo A Johnson & Susan P Holmes)
- In Nature, There Is Only Diversity (Michael R. McLaren, Benjamin J. Callahan)
- Consistent and correctable bias in metagenomic sequencing experiments (Michael R McLaren, Amy D Willis, Benjamin J Callahan)
Music: Eric Skiff — Come and Find Me (modified, licensed under CC BY 4.0).
Subscribe to the bioinformatics chat on Apple Podcasts, Pocket Casts, Spotify, or any other podcasting app via the RSS feed link. You can also follow the podcast on Mastodon and Twitter and support it on Patreon.
