Most Permissive Boolean Networks with Loïc Paulevé (#49)
August 19, 2020
In systems biology, Boolean networks are a way to model interactions such as gene regulation or cell signaling. The standard interpretations of Boolean networks are the synchronous, asynchronous, and fully asynchronous semantics.
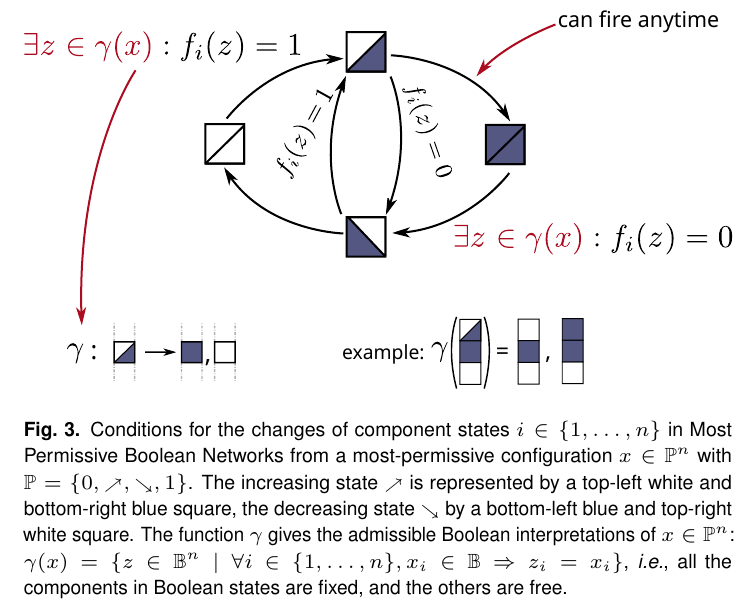
In this episode, Loïc Paulevé explains how the same Boolean networks can be interpreted in a new, “most permissive” way. Loïc proved mathematically that his semantics can reproduce all behaviors achievable by a compatible quantitative model, whereas the traditional interpretations in general cannot. Furthermore, it turns out that deciding whether a certain state in a Boolean network is reachable can be done much more efficiently in MPBNs than in the traditional interpretations.
Links:
- Reconciling Qualitative, Abstract, and Scalable Modeling of Biological Networks (Loïc Paulevé, Juraj Kolčák, Thomas Chatain, Stefan Haar)
- mpbn on GitHub: an implementation of reachability and attractor analysis in Most Permissive Boolean Networks
- BoNesis on GitHub: synthesis of Most Permissive Boolean Networks from network architecture and dynamical properties
Music: Eric Skiff — Come and Find Me (modified, licensed under CC BY 4.0).
Subscribe to the bioinformatics chat on Apple Podcasts, Pocket Casts, Spotify, or any other podcasting app via the RSS feed link. You can also follow the podcast on Mastodon and Twitter and support it on Patreon.
