Haplotype-aware genotyping from long reads with Trevor Pesout (#29)
January 27, 2019
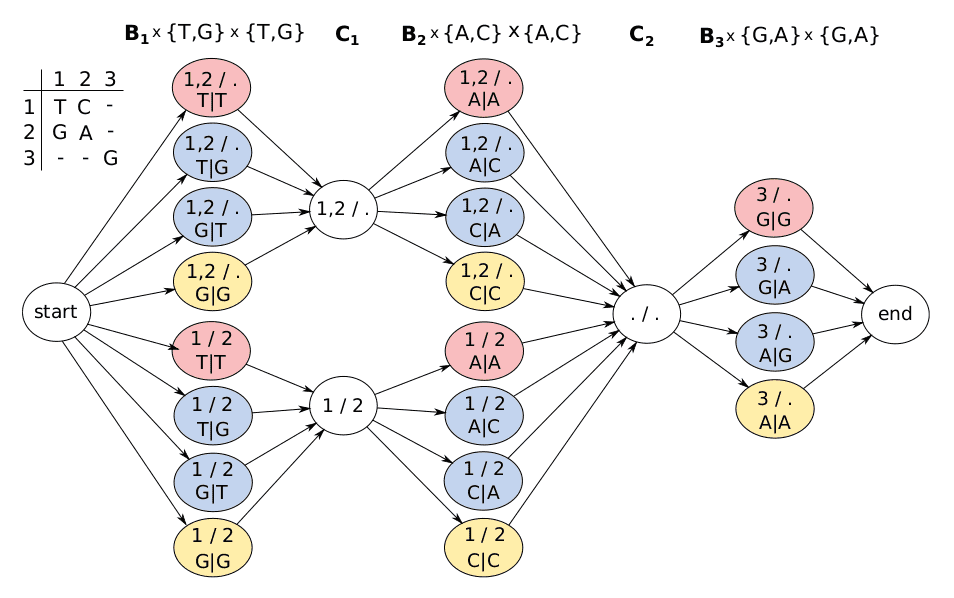
Long read sequencing technologies, such as Oxford Nanopore and PacBio, produce reads from thousands to a million base pairs in length, at the cost of the increased error rate. Trevor Pesout describes how he and his colleagues leverage long reads for simultaneous variant calling/genotyping and phasing. This is possible thanks to a clever use of a hidden Markov model, and two different algorithms based on this model are now implemented in the MarginPhase and WhatsHap tools.
Links:
Music: Eric Skiff — Come and Find Me (modified, licensed under CC BY 4.0).
Subscribe to the bioinformatics chat on Apple Podcasts, Pocket Casts, Spotify, or any other podcasting app via the RSS feed link. You can also follow the podcast on Mastodon and Twitter and support it on Patreon.
