scVI with Romain Lopez and Gabriel Misrachi (#36)
August 30, 2019
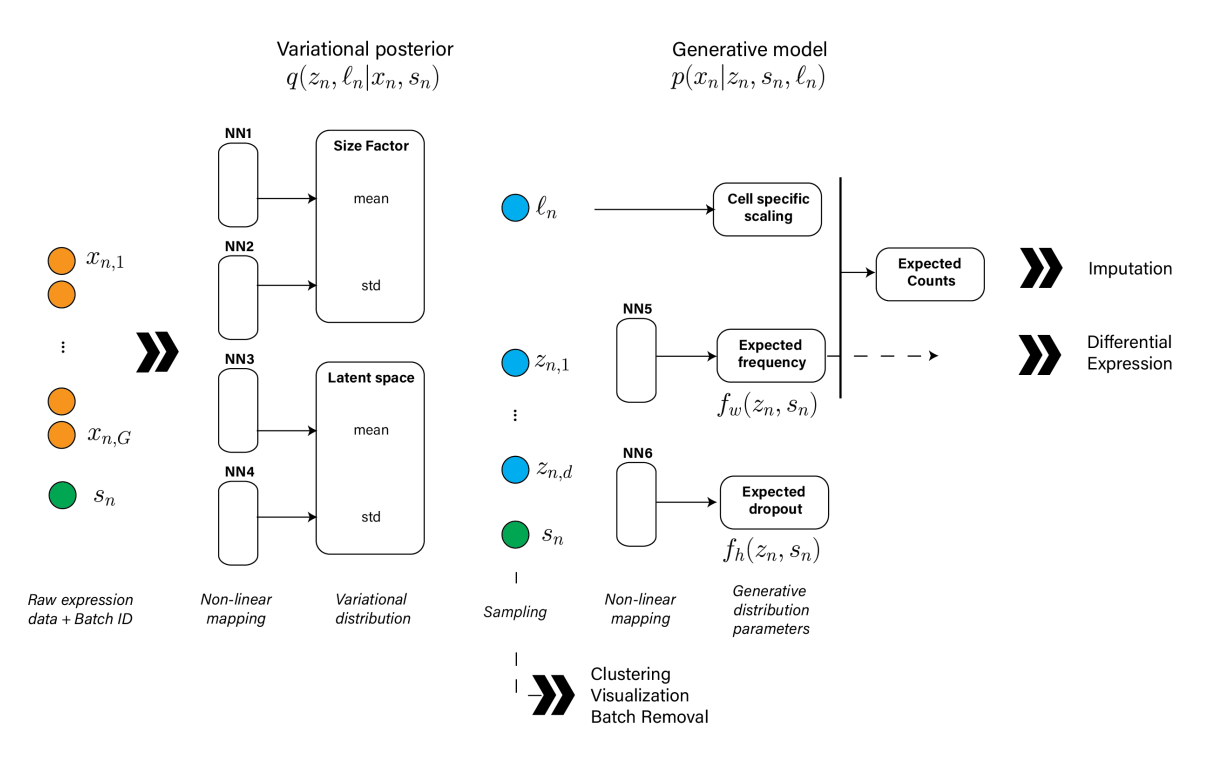
In this episode, we hear from Romain Lopez and Gabriel Misrachi about scVI—Single-cell Variational Inference. scVI is a probabilistic model for single-cell gene expression data that combines a hierarchical Bayesian model with deep neural networks encoding the conditional distributions. scVI scales to over one million cells and can be used for scRNA-seq normalization and batch effect removal, dimensionality reduction, visualization, and differential expression. We also discuss the recently implemented in scVI automatic hyperparameter selection via Bayesian optimization.
Links:
- Deep generative modeling for single-cell transcriptomics (Romain Lopez, Jeffrey Regier, Michael Cole, Michael I. Jordan, Nir Yosef)
- scVI on GitHub
- Should we zero-inflate scVI?
- Hyperparameter search for scVI
- Droplet scRNA-seq is not zero inflated (Valentine Svensson)
Music: Eric Skiff — Come and Find Me (modified, licensed under CC BY 4.0).
Subscribe to the bioinformatics chat on Apple Podcasts, Pocket Casts, Spotify, or any other podcasting app via the RSS feed link. You can also follow the podcast on Mastodon and Twitter and support it on Patreon.
